Page - Blog Single Column Right Sidebar
Current Research Projects
Understanding interactions in the human gut microbiome using synthetic microbial communities
Synthetic microbial communities constructed using pure isolates from the human gut are becoming popular vehicles to explore mechanisms of strain interactions, rules of community assembly, and to develop defined synthetic gut communities as potential therapeutics. Leveraging a collection of 100+ pure isolates representative of the human gut, a database of closed genomes derived from this collection of isolates, and the ability to perform high-throughput multi-omics measurements on microbial communities, we attempt to investigate strain-strain and strain-nutrient interactions within a complex but defined microbial community constructed from individual isolates (synthetic community). We perform in vitro growth experiments of synthetic communities to understand abundance and metabolic changes due to precise manipulations of strain presence and nutrient availability via defined growth media. In addition, germ free mice humanized by synthetic communities illustrate community transformation due to host interactions that may be overlooked in in vitro experiments. Together, we hope to gain predictive models enabling control of community composition and metabolic output.
High-throughput platforms for human microbiome research
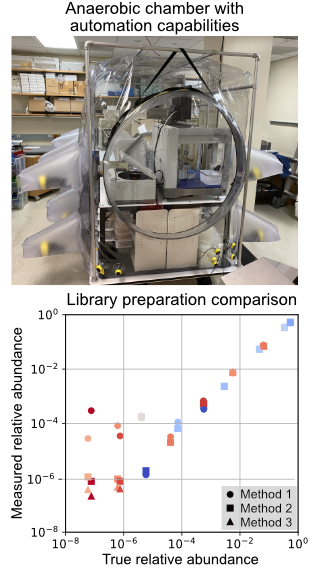
Human microbiome research relies upon DNA sequencing technologies to measure strain level relative abundance within a community and to reveal genes and pathways responsible for mechanisms of nutrient metabolism. The increasing scales of human, mice, and synthetic microbiome experiments require improvement in the experimental and analytical technologies with the objective to increase throughput and to lower financial barriers. The project to build high-throughput platforms for microbiome research focuses on several aspects of the research process:
1. Construct automation capabilities in a large anaerobic environment for the cultivation and phenotypic measurement of microbial isolates and communities.
2. Optimize and benchmark high-throughput and low-cost sample preparation pipelines for Illumina sequencing, using common samples generated from microbiome research including isolates, defined communities, and undefined communities.
3. Close genomes de novo for microbial isolates obtained from the human gut using long read technologies such as Oxford Nanopore.
4. Build AWS-based computational tools to analyse metagenomic sequencing data in a strain-aware fashion.
Past Research Projects
Microfluidic-based mini-metagenomics for discovering novel microbes from the environment
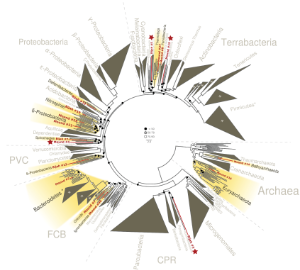
Microbes are important in global nutrient cycles and many aspects of our lives. Due to difficulties in cultivating pure species in lab conditions, much of the environmental microbial diversity has not been characterized. Recent reduction in DNA sequencing cost gave rise to popularity in metagenomics, a method to investigate unknown microbial diversity and constructing microbial genomes by directly sequencing genomic DNA recovered from environmental samples.
In this project, we developed a microfluidic-based mini-metagenomic approach that improves the identification of novel phylogenetic groups. This approach uses microfluidic parallelization to separate an environmental sample into many small sub-samples containing a few cells, significantly reducing complexity of each sub-sample and allowing high quality assembly while enabling higher throughput than typical single-cell methods. Although each sub-sample contains a limited mixture of several genomes, single-cell resolution is regained through correlations of genome co-occurrence across sub-samples, which in turn enables a rigorous statistical interpretation of confidence and genome association. As a result, the mini-metagenomic method combines advantages of current shotgun metagenomic and single-cell approaches. Our mini-metagenomic method is implemented on a commercially available Fluidigm© C1 microfluidic platform, which leverage the advantages of automated fluid handling. In addition, statistics derived from co-occurrence patterns enable statistical confidence in associating contigs (bioinformatically assembled DNA sequences) into genomes.
Applying the method to two hot spring sample from Yellowstone National Park, we successfully revealed genomes belonging to multiple known and novel bacterial and archaeal lineages. Functional analysis revealed that these organisms utilize diverse pathways for energy metabolism. The resolution of the mini-metagenomic method enabled accurate quantification of genome abundance, even for rare genomes. Our analyses also revealed a wide range of genome level single nucleotide polymorphism (SNP) distributions. These results demonstrate the power of our microfluidic-based mini-metagenomic approach for discovering novel phylogenies. We foresee the method to be broadly applicable to answer questions related to environmental microbial communities.
Investigating cyanobacterial physiology using microfluidics
Cyanobacteria are important agents in global carbon and nitrogen cycling and hold great promise for biotechnological applications. Genetically tractable model organisms such as Synechocystis sp. and Synechococcus sp. have advanced our understanding of photosynthetic capacity and circadian behavior, mostly using population-level measurements in which the behavior of individuals and population heterogeneity cannot be monitored. Therefore, a single-cell level understanding of the phenotypic heterogeneity across populations and how environmental factors such as changes in light affect growth may provide insight into how cells integrate external stimuli with internal mechanisms of cell-cycle and cell-size regulation. This understanding may also be required for optimizing the efficiency of large-scale Synechocystis bioreactors.
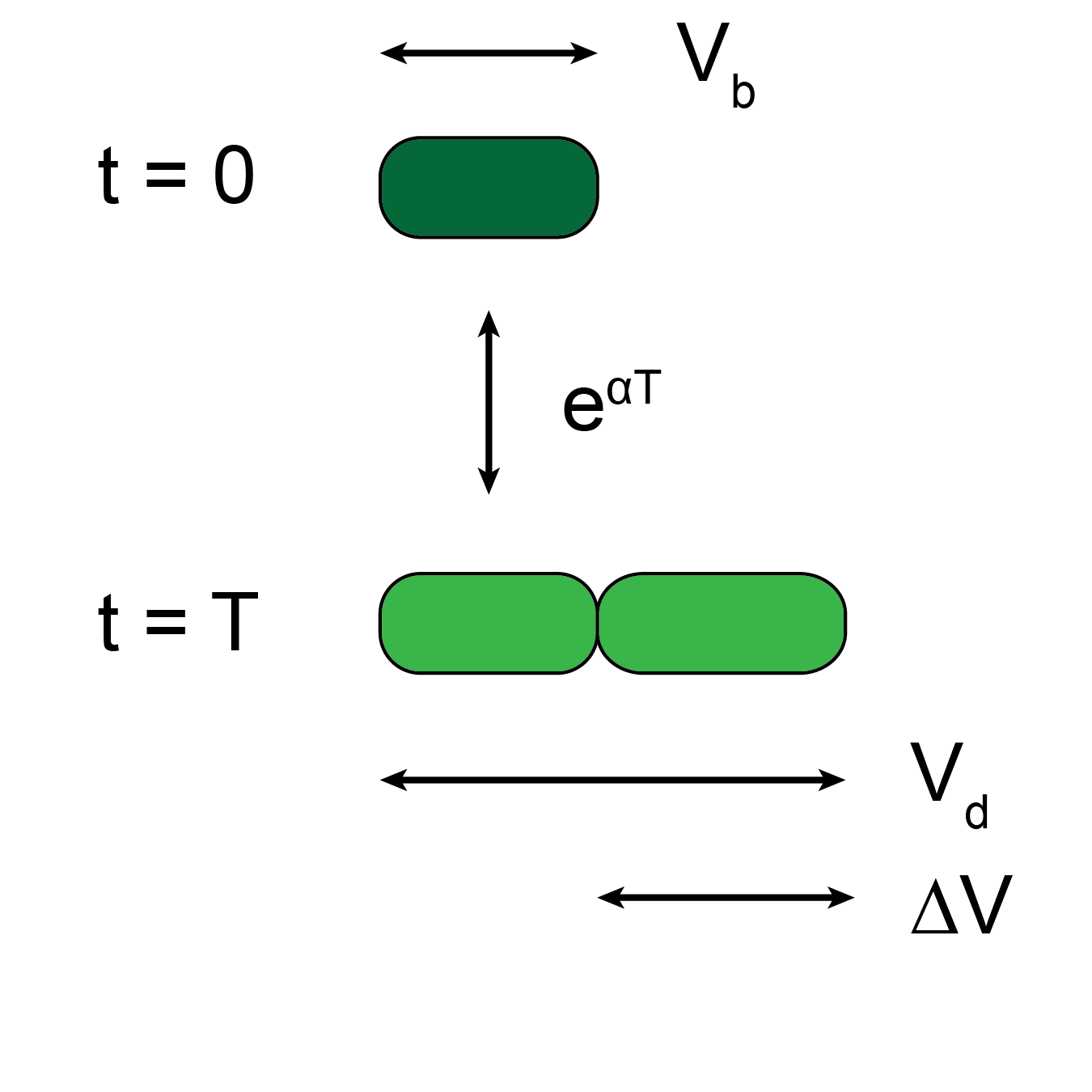
Tracking Synechocystis cells at the single-cell resolution is challenging due to their morphology and slow growth rate. To overcome this challenge, we modified a microfluidic cell-culture system and developed an image analysis pipeline to monitor Synechocystis growth and division over several generations in continuous illumination or with light-dark cycling. Using parameters extracted from single-cell growth curves, we saw that cells expand their volume exponentially. When exposed to light-dark cycles, growth and division only occurred during the light period. Although dark periods halt growth and division, they do not disrupt cell-cycle control. Finally, relationships between birth size, division size, and generation time are compared to simulation results to reveal the most consistent model of cell-cycle regulation. We believe that our microfluidic setup and image analysis pipeline can be used to explore phenotype of a much larger variety of bacterial species and answer questions related to bacterial adaptations to environmental changes.
Post-fabrication testing of multilayer microfluidic devices
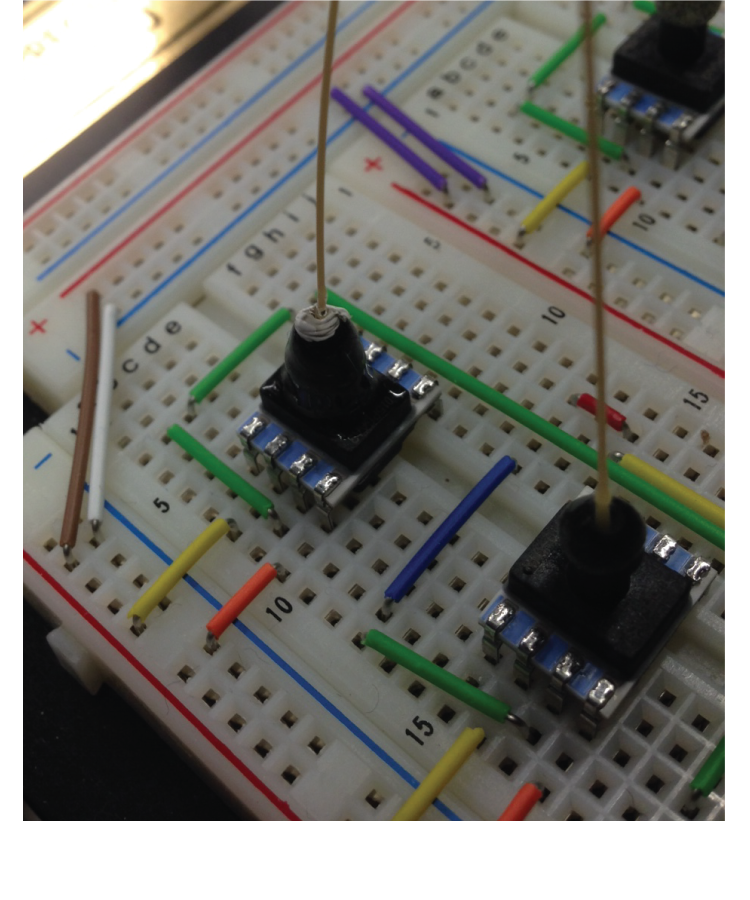
Because of its small reaction volume, high throughput capability, and amenability to automation, microfluidic technology has seen tremendous growth in biological, chemical, and medical fields. Multilayer, pressure driven, PDMS (polydimethylsiloxane) microfluidic devices, in particular, have been commercialized in many applications. Similar to an electronic transistor, the microfluidic valve developed in the Quake Lab acts as a switch directing fluid flow on-chip. This control component allows for large scale integration of hundreds of individual reaction components. However, as the complexity of microfluidic devices increase, yield decreases, especially for custom chips made in research labs. It is often difficult to identify defects in complex microfluidic chips by eye, leading to significantly longer design-build-test cycles.
Borrowing concepts from electronic post-fabrication testing, we constructed a setup to perform automated post-fabrication testing of multilayer microfluidic devices. Collaborating with the Chakrabarty group from Duke University, we generated circuit models of microfluidic devices including functional effect of different design and fabrication errors. Using test patterns specifically designed to target functional defects, we demonstrated experimentally the applicability of using electronic post-fabrication algorithms to detect functional errors in microfluidic devices. We hope that the use of automated testing in the fabrication process will decrease the design-build-test cycles and lower the barrier of using microfluidic technologies.
Microfluidic serial digital to analog pressure converter
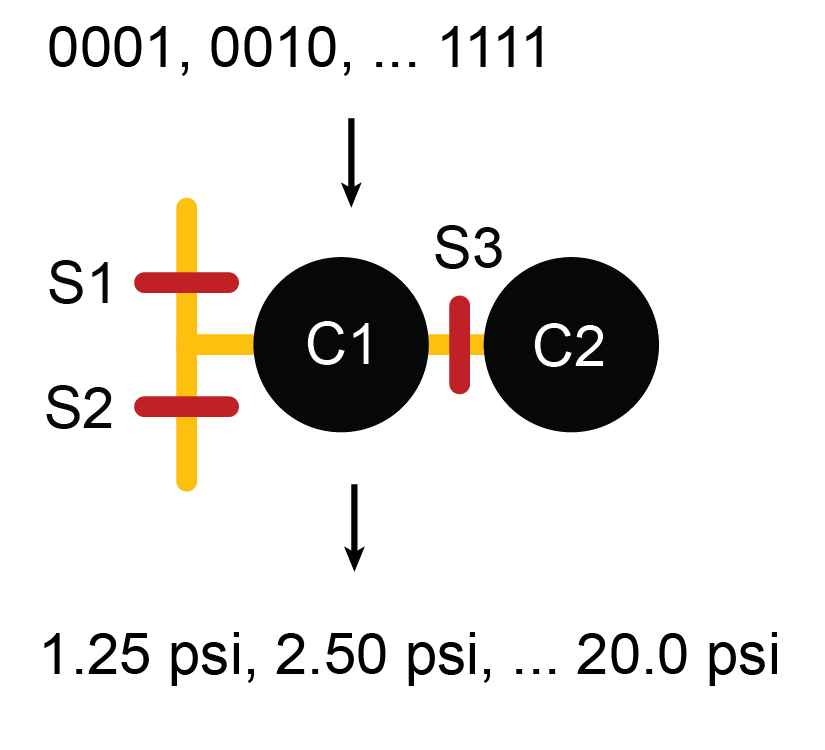
Multilayer microfluidics based on PDMS (polydimethylsiloxane) soft lithography have offered parallelism and integration for biological and chemical sciences, where reduction in reaction volume and consistency of controlled variables across experiments translate into reduced cost, increased quantity and quality of data. Even though microfluidics, using push up or push down valves, offer great power for precision measurement and analysis, they do have limitations. One such limitation is that the on-chip elastomeric valves can only act like digital switches, allowing or preventing flow. The same valve cannot provide a range of analog actuation pressures to partially restrict a channel without requiring the user to manually adjust bulky, external pressure sources. When it is necessary to vary pressure and flow rates of multiple flows on the same device, an on-chip method to create a range of pressures from a single, constant, external pressure source is preferred.
In this project, we designed and fabricated a microfluidic serial DAC (Digital to Analog Converter) that can be integrated with any PDMS device to expand its functionality, effectively adding an on-chip pressure regulator. The microfluidic serial DAC can be used with any incompressible fluids and operates in a similar fashion as an electronic serial DAC. In this device, PDMS diaphragms act as fluidic capacitors to store pressure. Experimental results demonstrated linear transfer function with a 4 bit binary code, resulting in 16 distinct pressure levels. In addition, this component has a maximum refresh frequency of 3 Hz and can maintain pressure for up to 10 minutes. Therefore, the microfluidic serial DAC adds to the repertoire of versatile on-chip control elements.
Fabrication of MEMS check valves

Parylene is a popular material used in implantable devices due to its flexibility and biocompatibility. In the MEMS lab at Caltech, parylene-based MEMS (microelectromechanical system) check valves are used in implantable and in vitro devices to regulate pressure and flow of liquids. The property differentiating various passive check valves is their cracking pressure, defined as the lowest pressure at which flow is permitted.
In order to control cracking pressure of fabricated parylene check valves, we explored two separate approaches. Since materials differ in surface properties, coatings may change surface adhesion (stiction) with thin parylene films, resulting in different cracking pressures. To test this idea, we subjected silicon substrates to various treatments, metal coatings and found that by adjusting surface treatments, a large range of cracking pressures can be achieved. Another direction involved using gray scale photolithography to pattern slanted photoresist profiles. Parylene is then deposited on top of the photoresist as a thin film. After fabrication, thermal annealing is performed to increase tensile strength in the slanted parylene tethers. In this process, the time and temperature of the annealing process are tuned to achieve the most desirable cracking pressure.